Guest post by Mónica Arrúe and Gorka Epelde from Vicomtech
In the context of MIDAS project, a data preparation sync functionality has been implemented to allow policy site data owners to prepare data in different iterations and automatically deploy prepared data to MIDAS platform.
This functionality has been implemented in GYDRA; the evolution of the TAQIH data preparation tool, which used a single machine focussed development (with machine memory fitting dataset size limitation), to a more distributed approach. This was motivated by policy members shared large datasets and Big Data (BD) nature of the MIDAS project. In the following picture the data preparation sync workflow is represented.
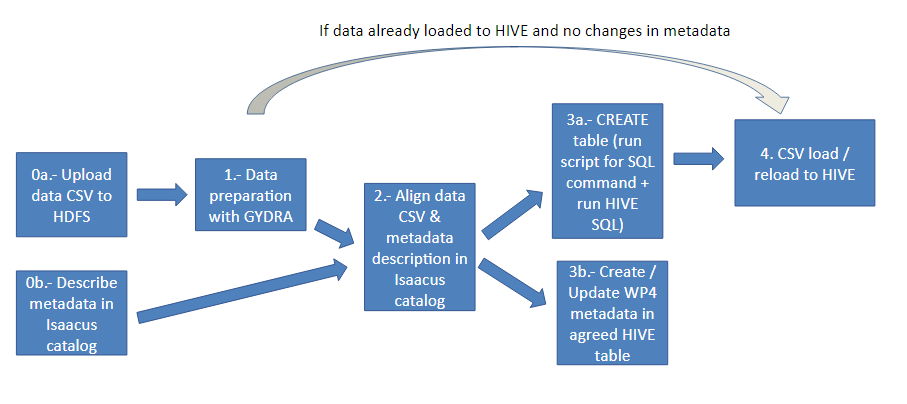
Figure 1. Data preparation sync flow
Firstly, data .csv files are uploaded to HDFS (step 0a) from their corresponding virtual machines. HDFS (Hadoop Distributed File System) is a data stored system which implements a distributed file system that provides a high-performance access to data. Meanwhile, metadata is described in Isaacus metadata catalogue (step 0b). Once the data is loaded in HDFS and its metadata described in Isaacus, dataset needs to be registered in the GYDRA interface by filling certain information, including Isaacus study and dataset reference, as it can be seen in the following picture.
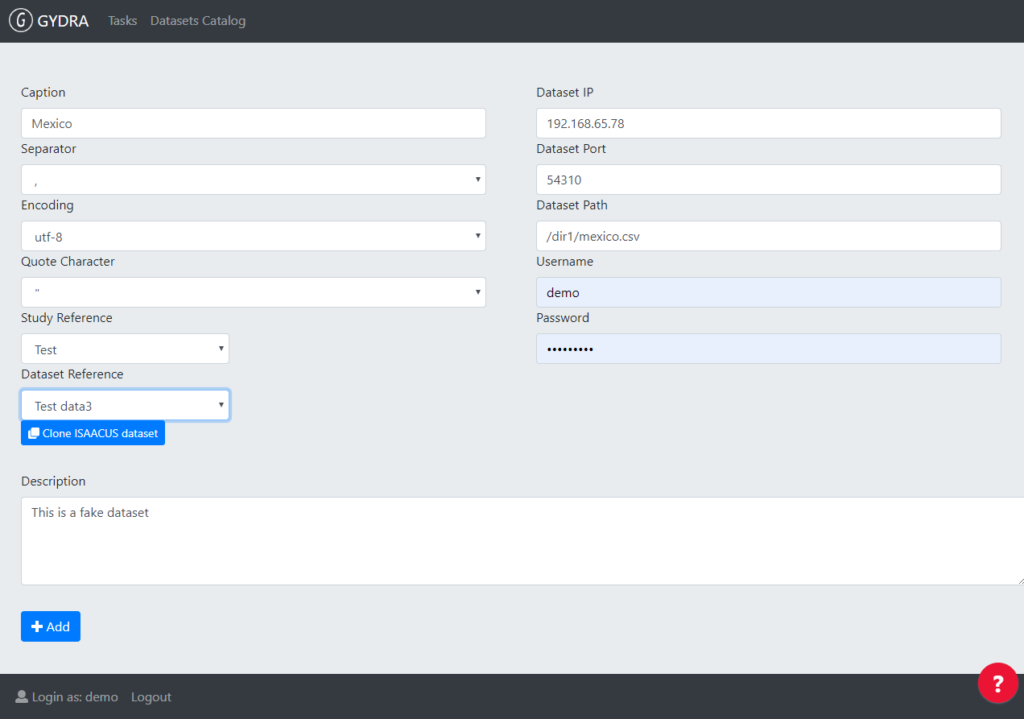
Figure 2. Dataset registration UI in GYDRA. Isaacus study and datasets references are filled here.
Once the dataset is registered in the GYDRA database, the user can see the registered dataset in the interface of the dataset catalogue, can trigger pre-processing of it with the GYDRA tool button and can prepare it in GYDRA (step 1).
Upon a data preparation phase, a dataset and its metadata alignment phase must be launched (step 2). The aim of the alignment phase is to make sure that the metadata described by data owners in the Isaacus server matches the metadata inferred by the GYDRA tool’s pre-processing (i.e. pre-processing of the result of data preparation). This alignment is needed since many of the GYDRA to MIDAS platform deployment scripts rely on Isaacus metadata to feed the required data preparation sync scripts. In the following image a list of datasets registered in GYDRA tool and the buttons to trigger the datasets alignment marked in red can be seen.
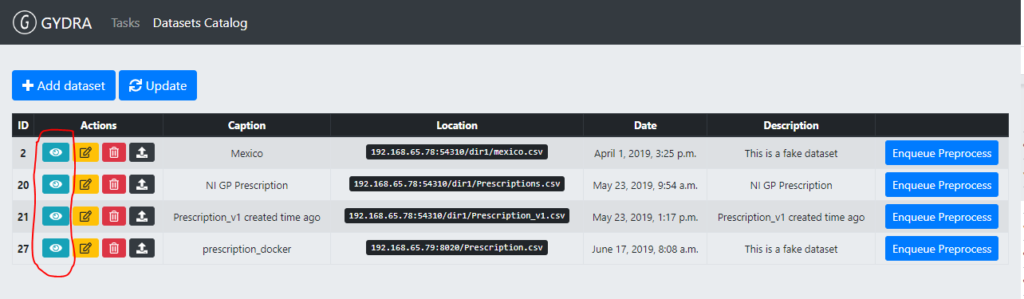
Figure 3. Dataset alignment trigger in the GYDRA user interface
The GYDRA pre-processing automatically detected variable data types may not match those stored in the Isaacus server. In addition, since the process of incorporating metadata into the Isaacus application is manual, variable names and other descriptions are likely to contain errors. Consequently, the metadata alignment tool has been developed within the GYDRA tool to detect data source and metadata mismatches (coming from data owners) and to identify human errors which occurred during Isaacus metadata names and data types definition, and to detect accidental variable name / data types modifications made during data preparation (step 1). The image below shows an example of a dataset metadata alignment interface.
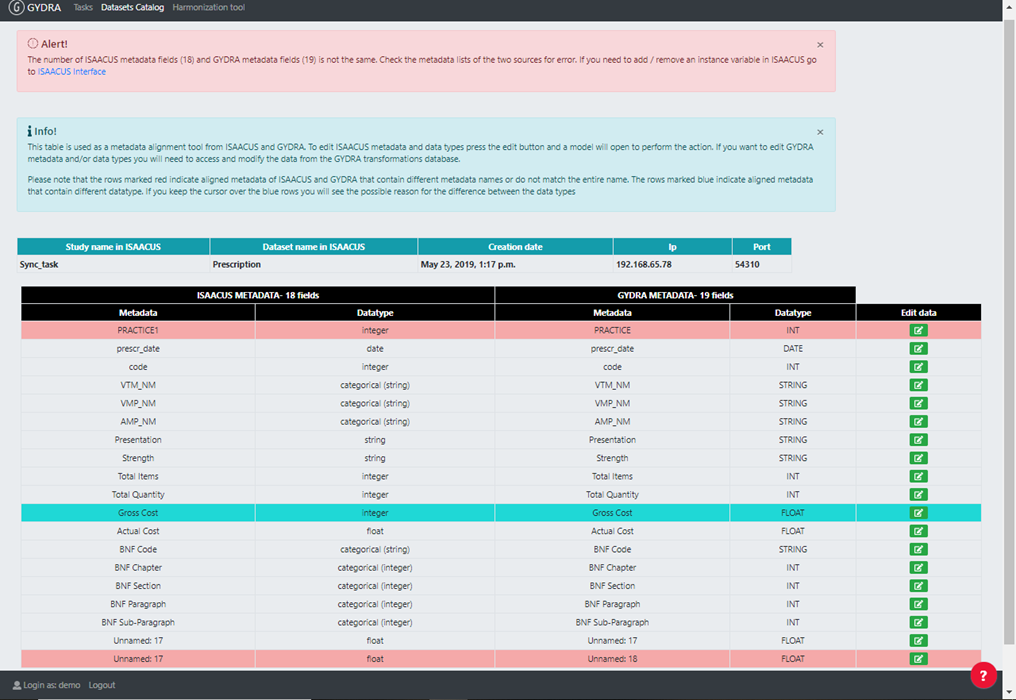
Figure 4. GYDRA application metadata alignment interface
To create the alignment tool a technique has been used to match metadata names that are lexically equal or similar, since the objective of the alignment tool is to align the names of the variables from both sources that most resemble.
Among the techniques based on lexical coincidence the Levenshtein distance has been applied. Levenshtein calculates the smallest number of operations or changes that must be made in a word or a set of words to convert it into another word or into another group of words. It is a very used and simple technique to find coincidences between the names of the variables. The Levenshtein distance has been applied to the variable names of both sources, aligning the variable names. In the table the data types have been included next to the variable names.
Once the table is created, red and blue colours have been used so that the user can easily detect changes or variations in the names of variables and/or data types. On the one hand, the lines in which the names of the aligned variables do not coincide are highlighted red. On the other hand, blue colour is used to indicate that the data types of the aligned variables are not the same. In addition, if the user keeps the mouse of the keyboard on the blue line, the possible reason for the difference between the two types of data will display in a comment.
Pressing the button in the edit column (right-hand column from Figure 4) opens a window that allows to change the name and/or data type of the variable stored in Isaacus. However, it does not allow modifying the GYDRA metadata. This requires going through GYDRA transformations to get variables corrected on the real data.
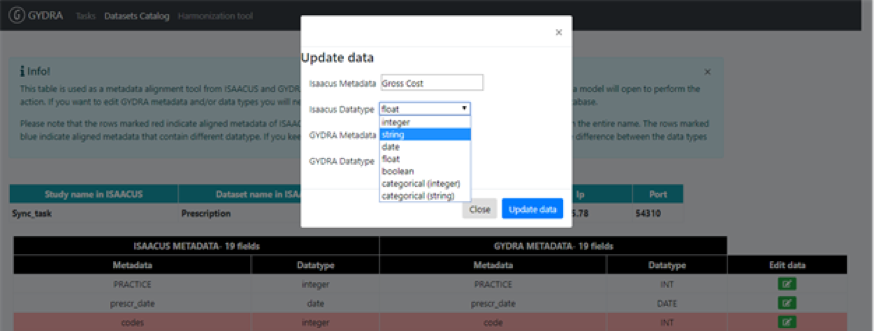
Figure 5. The modal window that opens when “Edit” button of a line is pressed on the metadata alignment table in GYDRA interface.
After GYDRA data and Isaacus metadata are aligned, prepared data can be deployed to MIDAS platform (steps 3 and 4). To trigger the deployment of a dataset, the user needs to click on “Deploy dataset” button in the GYDRA registered dataset list (Figure 6).
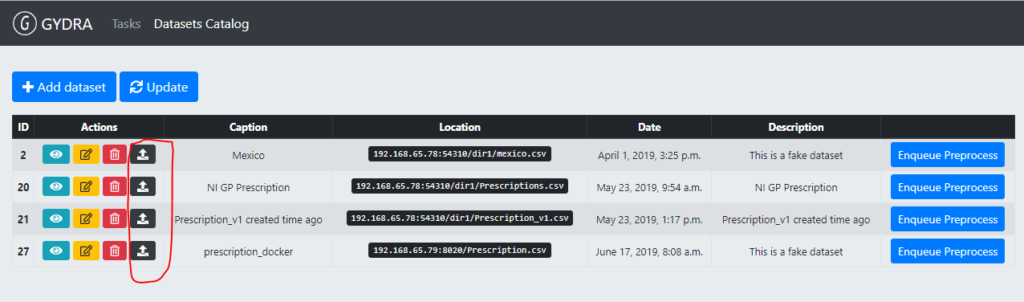
Figure 6. Dataset deployment trigger in the GYDRA user interface
On the one hand, a script is executed to create the data table in HIVE taking as reference the information of the updated data from GYDRA (i.e. variables’ names and data types) (step 3a) and the data is collected from and loaded into this table (step 4). This script generates a new database in HIVE with the name of the study in Isaacus metadata where the created table is stored. On the other hand, the metadata (named as WP4 metadata in MIDAS) table that enables to offer and limit the analytics and visualisations available in the MIDAS platform, is generated and uploaded to a database in HIVE called “wp4metadata” (step 3b). The information to generate this table is retrieved from Isaacus metadata.
If data prepared with GYDRA were already uploaded to HIVE and WP4 metadata as well, and in the data preparation there has been no changes in the metadata, it only reloads the data back into the table already created in HIVE.
In addition, code dictionaries of categorical variables which are described in Isaacus are retrieved from there and tables are created in HIVE, more specifically in a database called “dictionaries”. These tables have two columns: “id”, which stores categorical variables’ code, and “description” that stores these codes’ associated value, as it can be seen in the image below.
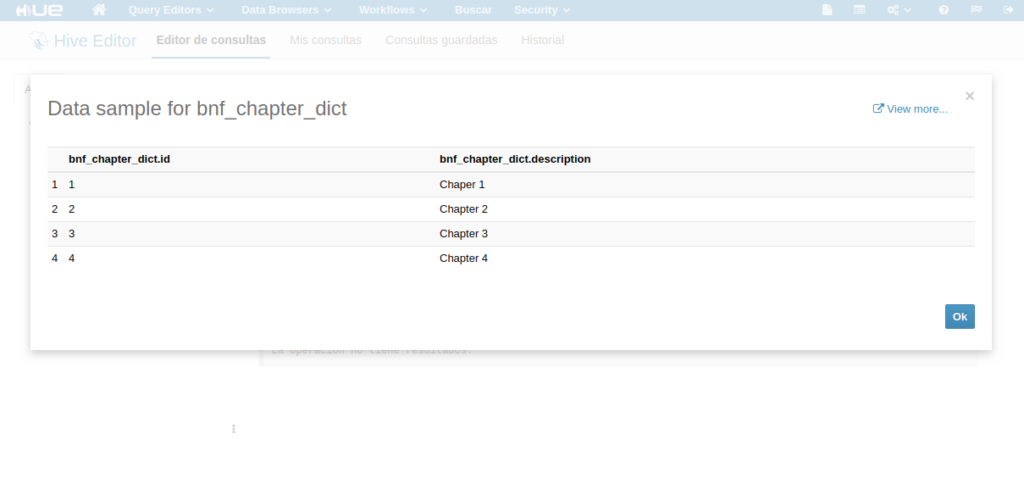
Figure 7. Example of codes dictionary table in HIVE
An example of the result of deployment of an open data dataset called “prescription” on the MIDAS platform is shown next. Firstly, it can be seen in the following image that three new databases have been created in HIVE: “dictionaries”, “sync_task” (sync_task is the name of the study in Isaacus under which prescription dataset is described) and “wp4_metadata”.
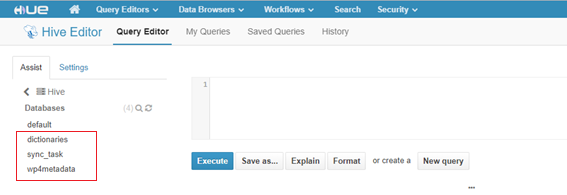
Figure 8. Deployment of a dataset on the MIDAS platform. Shows the data preparation sync generated Hive databases (i.e. dictionaries, sync_task and wp4metadata)
In the following image, the created table within “sync_task” database is shown, which contains the prepared data of the deployed table.
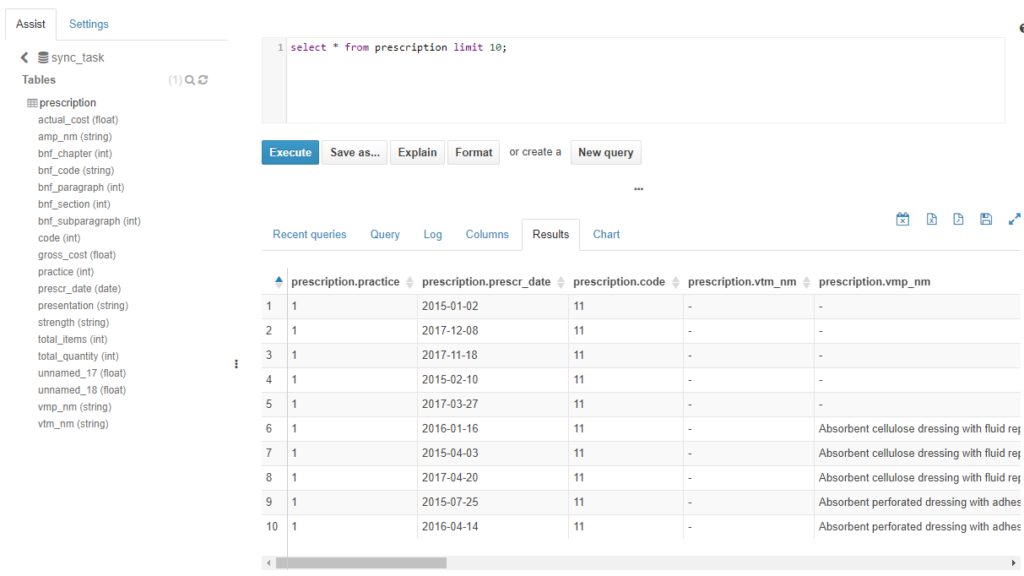
Figure 9. Deployment of a dataset on the MIDAS platform. Shows “prescription” table created in Hive and prepared data loaded into it
On the other hand, “dictionaries” database contains a table called “bnf_chapter_dict”, which has the categorical dictionary of the “prescription” table.
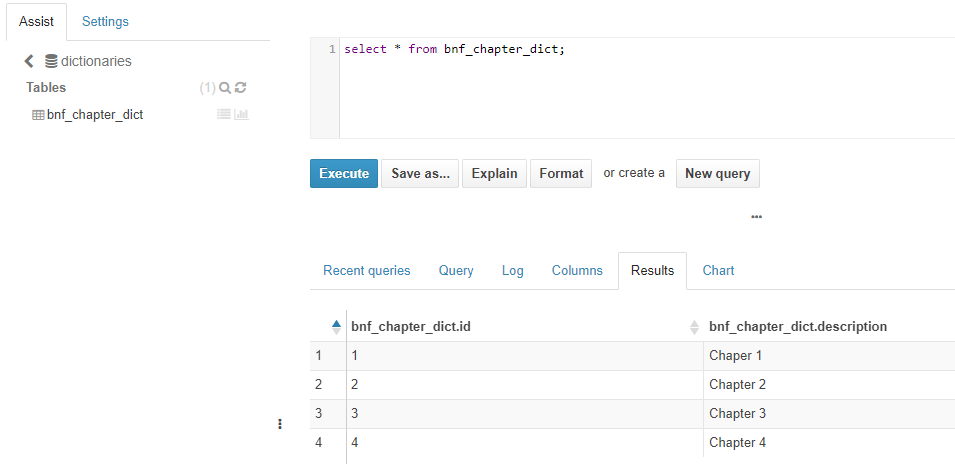
Figure 10. Deployment of a dataset on the MIDAS platform. Shows bnf_chapter_dict categorical variables’ generated Hive table (within Hive dictionaries database) and the dictionary content loaded into it
Finally, WP4 metadata database contains the table “wp4_metadata” created needed to create analytics of the MIDAS platform, as explained before.
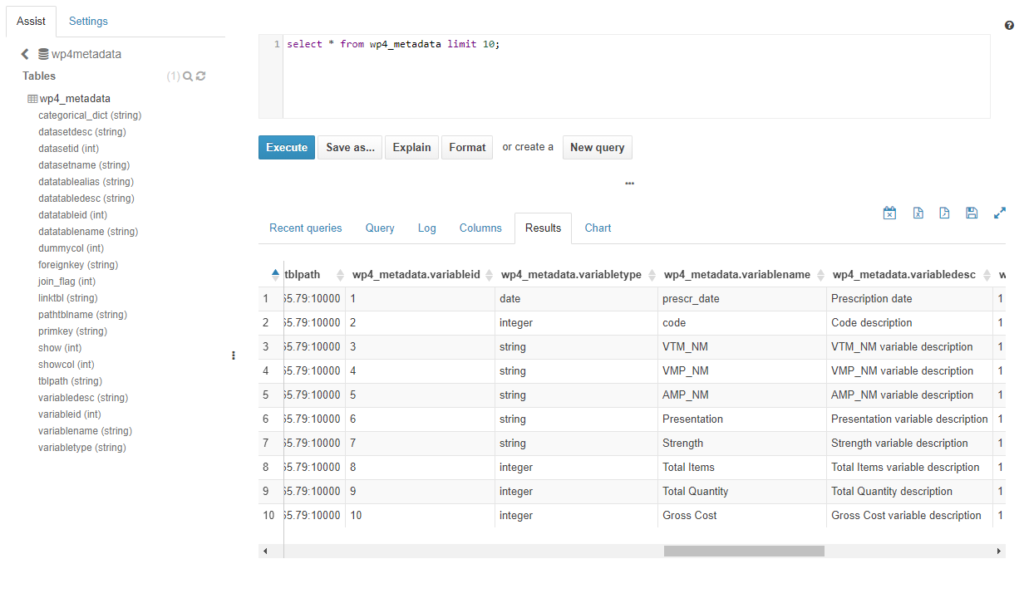
Figure 11. Deployment of a dataset on the MIDAS platform. Shows Hive table generated for WP4 metadata (within Hive wp4metadata database) and the metadata content loaded into it
