Guest post by Gorka Epelde (Vicomtech) and Jaakko Lähteenmäki (VTT)
In a previous blog post, we introduced the data ingestion methodology for individual datasets, and shortly introduced datasets description with the ISAACUS GSIM-based metadata model to facilitate the identification of harmonisable attributes and the subsequent harmonisation of the datasets. In this blogpost we describe more in detail the adopted common metadata model and data harmonisation approach.
A common data model is needed in the MIDAS project in order to enable automatic processing of data sets, enable reuse of data analytics components, run cross-site analytics and enable common understanding of the data contents via metadata.
Many of the existing data set models in the state of art are focused to a certain purpose (e.g. SCDM, OMOP or PCorNet), for example addressing specifically the collection of clinical trial data or the comparison of health care provider efficacy. Such models are relevant concerning the needs of the MIDAS project, but do not provide a holistic solution for the large variety of source datasets involved in MIDAS. Some of the models (e.g. GSIM, GSBPM, CSPA and SDMX) take a more generic approach and are therefore relevant concerning a common data model for the MIDAS project.
For MIDAS, we have opted for a common metadata model approach (ISAACUS GSIM-based metadata model) where datasets metadata are described and then a search of harmonisable variables and specific transformations are defined upon search results. This contrasts with the alternative approach of defining a (or adopting existing) common data model and transforming the source datasets to such data model.
MIDAS interoperability approach is built around the selected metadata model-based approach, including datasets cataloguing, variable annotation, harmonisable variable identification and scripts generation to automate transformed datasets reloading and use by the analytics platform.
Datasets cataloguing approach following ISAACUS metadata model
- Starting from the dataset description (described in the previous blog post) the ISAACUS metadata model required information is inserted to the ISAACUS server.
- Variables are annotated to enable the identification of harmonisable variables in a subsequent step. For each variable (named InstanceVariable in the ISAACUS metadata model) annotation, the following is done:
- Assigning to the variable (InstanceVariable in the ISAACUS metadata model) at least a concept variable or variable classification index (named Variable in the ISAACUS metadata model). Variables can correspond to one or more classification index terms. Based on the state of art analysis, the classification index selected for variable annotation is the Maelstrom Classification.
- Assigning topics (Adding ConceptScheme element to Concept in the ISAACUS metadata model), from platform loaded concept schemes (i.e. YSO, MeSH and TERO ontologies). This is reflected as keywords in the ISAACUS server tool.
Harmonisable variable identification strategy
The harmonisable variable identification strategy starts from the annotated classification index, looking at domains and subdomains from adopted Maelstrom Classification, and identifying where different data sources share elements. A tool is being developed to summarise per subdomain elements in a table and visualise comparative variable information per each classification item.
Based on this summary and per variable classification index information, ISAACUS server catalogue will be used to obtain further information on those variables. Additionally, the ISAACUS server’s search functionality allows to search among variables by name, description and keywords, to refine the identification of harmonisable variables from the summary or identify mis-classified variables that should be considered. The following figure shows the results of the ISAACUS server’s search functionality:
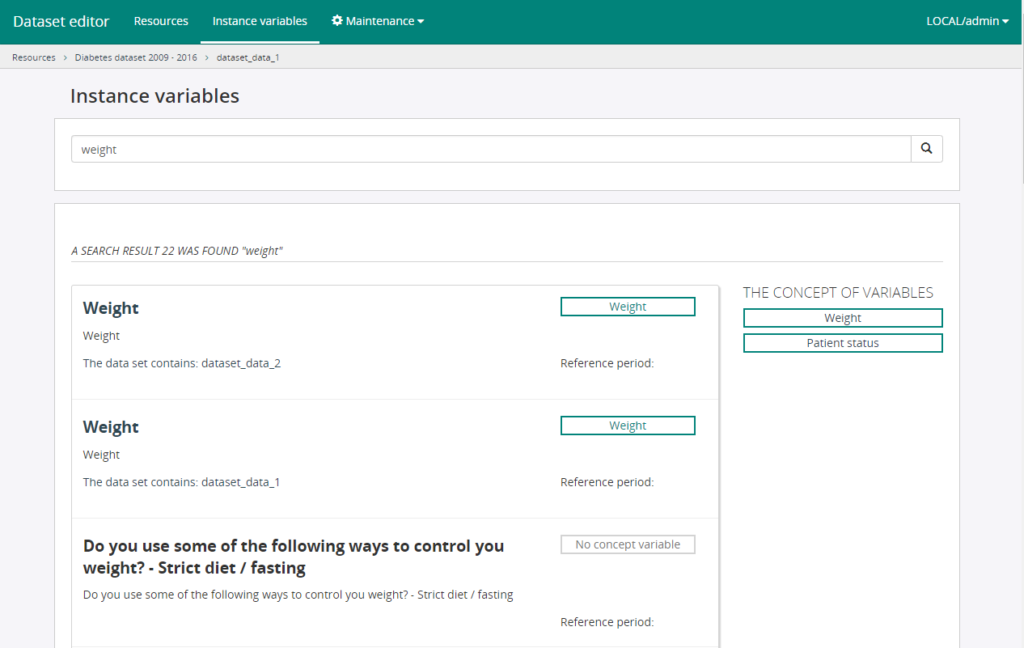
Figure 1. Results of the ISAACUS server’s search functionality.
From the search results of the ISAACUS server’s search functionality, we can also identify concept variables related to the search and identify mis-classifications or navigate to variables under certain classification index by clicking on the concept variable item. The following figure shows the resulting variables for a classification index:
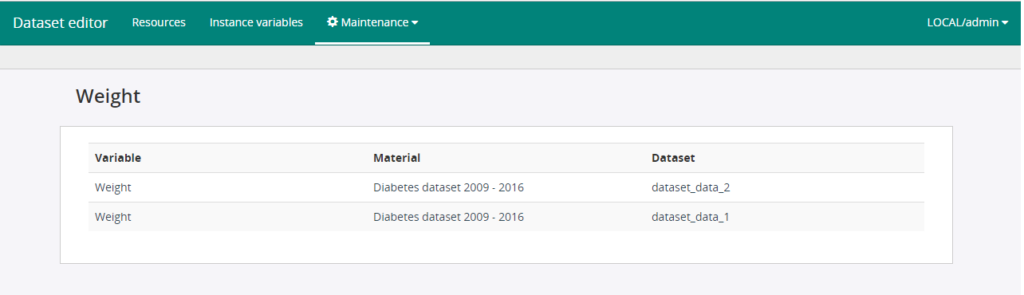
Figure 2. Resulting variables for a classification index.
Data transformation approach for dataset harmonisation
Upon identifying harmonisable variables across datasets and agreeing on a final set of variables and their units and codings, the corresponding metadata is inserted to the ISAACUS server and datasets are required to be transformed.
Despite different approaches were thought and tested, an alternative to use and extend MIDAS project developing GYDRA data preparation tool (renamed from TAQIH) has been selected.
Currently the GYDRA tool allows to define and run pipeline of dataset modification action targeting the quality improvement, which off-the-shelf supports data harmonisation tasks related data transformation to obtain a target dataset on CSV starting from a source CSV. Supported harmonisation functionalities include:
- Deleting a variable (GYDRA DROP feature)
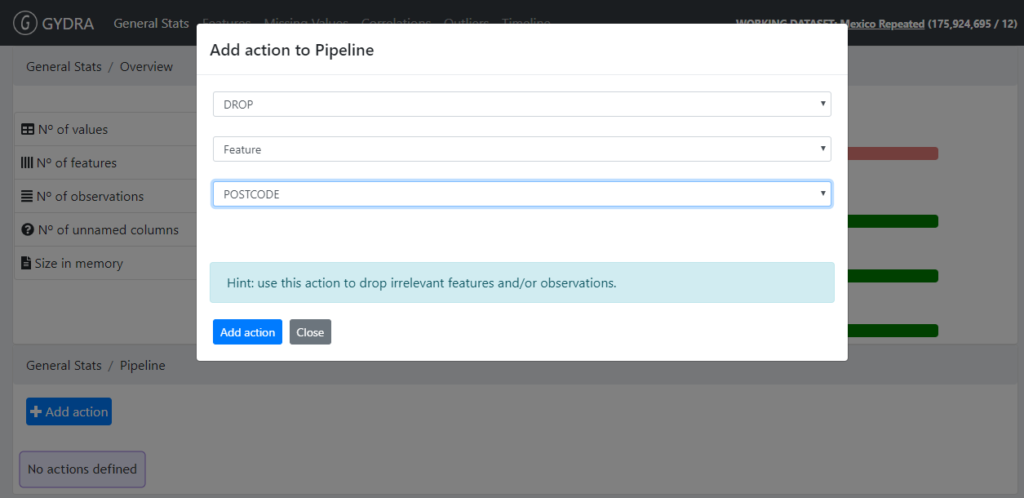
Figure 3. GYDRA DROP feature
- Renaming a variable (GYDRA RENAME feature)
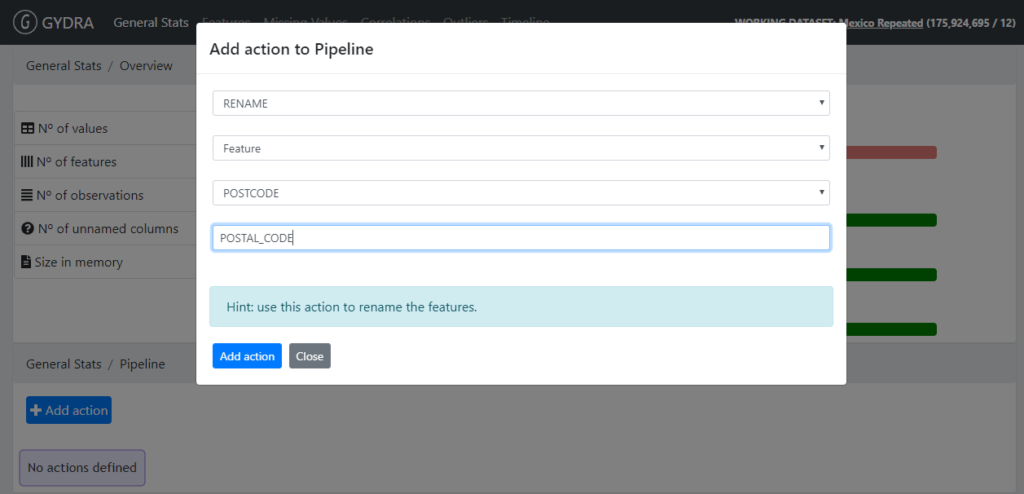
Figure 4. GYDRA RENAME feature
- Changing coding of categorical values (GYDRA CHANGE_VALUE)
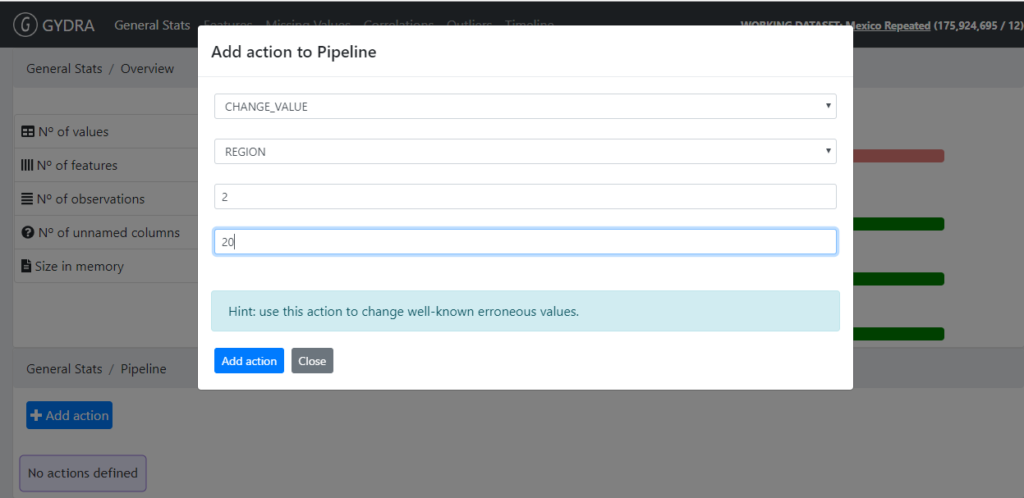
Figure 5. GYDRA CHANGE_VALUE feature
- Continuous unit changes (GYDRA OPERATE with add / subtract / divide / multiply operators)
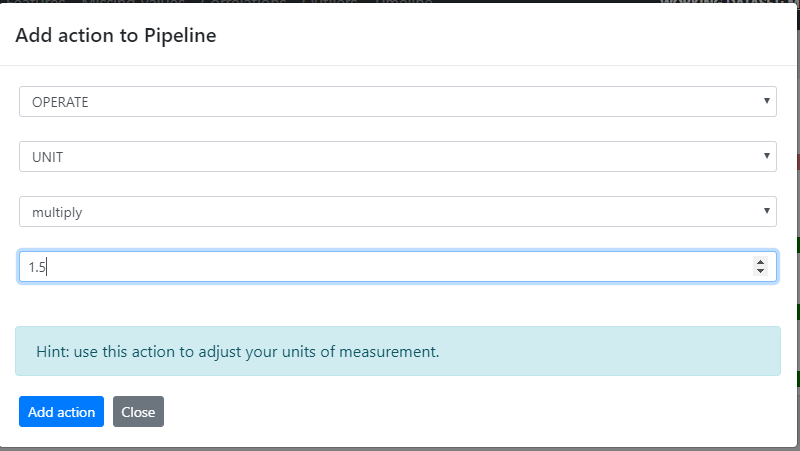
Figure 6. GYDRA OPERATE feature with add / subtract / divide / multiply operators
Despite many data transformations can be achieved with the existing GYDRA functionalities, new functionalities have been identified as operating over two variables to generate a new one (e.g. BMI value based on height and weight) and more complex transformations that will need to be targeted by integrating external services (e.g. transformation of clinical codes).
Additionally, GYDRA allows the user to define and update dataset transformation pipelines visually and in an interactive way (See Figure 7).
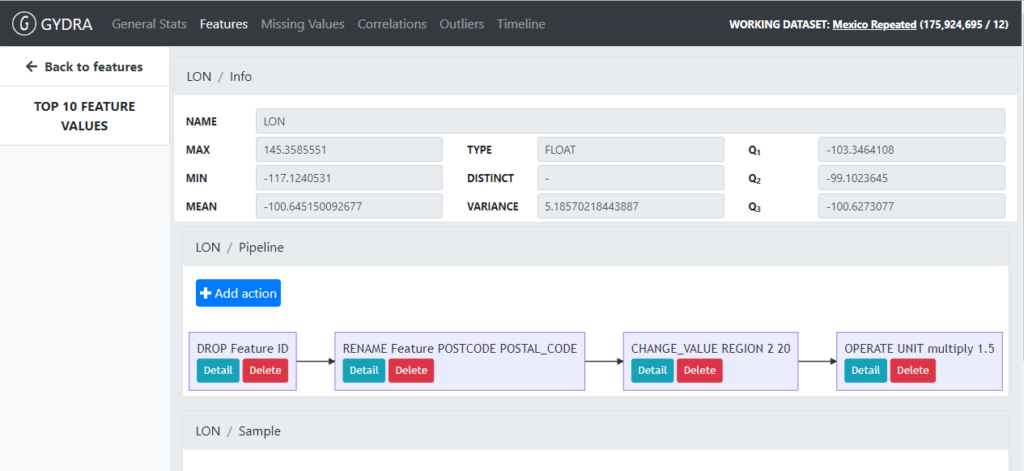
Figure 7. GYDRA visual and interactive dataset transformation pipeline definition
Having the metadata of the datasets described on the ISAACUS server, scripts have been generated to convert the described metadata into the format required by the analytics platform to feed MIDAS Dashboard with the required information. Additionally, scripts have been created to generate the required Hive table creation and data loading queries starting from the metadata described on the ISAACUS server. Through these scripts the resulting transformed dataset in CSV format, are placed in HDFS and loaded into Hive and ready for analytics and visualisation through the generated analytics platform metadata.
